I was talking to Dr. Brown yesterday, and he brought up an old nightmare of mine: removing prescriptions with negative quantity dispensed values.
He was explaining about how if a patient didn’t pick up their medications that the pharmacy would reverse the record or create a negative event. In other words, if my doctor prescribed me 30 tablets of Naproxen but I never picked them up, there would be a record for the 30 tablets, and then a record for the -30 tablets.
When I first started here, I wasn’t quite sure how to do it. I spent a lot of time trying and failing.

I called in Carl Henriksen for some help, and he got me on the right path.
But mind you, I was talking to Dr. Brown yesterday. I decided to revisit the issue myself and do it my own way.
Here was the issue we had (not the actual data – this is a reproducible example):
data have;
infile datalines dlm = " ";
length id $1. drug $10.;
input id drug service_dt :mmddyy10. qty_dspnd;
format service_dt mmddyy10.;
datalines;
1 Gabapentin 12/20/2015 -30
1 Gabapentin 12/20/2015 -30
1 Gabapentin 12/20/2015 30
1 Gabapentin 12/20/2015 30
1 Gabapentin 12/20/2015 360
1 Gabapentin 12/20/2015 30
1 Gabapentin 12/21/2015 30
1 Gabapentin 12/21/2015 30
2 Naproxen 12/27/2016 30
2 Naproxen 12/27/2016 -30
2 Naproxen 12/27/2016 30
3 Ibuprofen 12/28/2016 -30
3 Ibuprofen 12/28/2016 -30
3 Ibuprofen 12/28/2016 30
3 Ibuprofen 12/28/2016 30
3 Ibuprofen 12/28/2016 30
4 Naproxen 12/29/2016 30
4 Naproxen 12/29/2016 -30
;
And this is what we wanted:
data requested_want;
infile datalines dlm = " ";
length id $1. drug $10.;
input id drug service_dt :mmddyy10. qty_dspnd;
format service_dt mmddyy10.;
datalines;
1 Gabapentin 12/20/2015 360
1 Gabapentin 12/20/2015 30
1 Gabapentin 12/21/2015 30
1 Gabapentin 12/21/2015 30
2 Naproxen 12/27/2016 30
3 Ibuprofen 12/28/2016 30
;
Notice in our have dataset that we have some prescriptions on the same day that have negative values, while others have positive values. Some of those values are duplicated while some aren’t.

One step at a time.
We have negative and positive prescription values. Get out our old friend PROC SORT and do some sorting before we go any further.
proc sort
data = have;
by id drug service_dt qty_dspnd;
run;
I sorted by id, drug, service_dt, and qty_dspnd to get records ordered sequentially for the same day. Prescription quantites are in ascending order, with the lowest appearing first and the highest appearing last for a given id, drug, and service_dt. Here’s what we’re looking at now:
id drug service_dt qty_dspnd
1 Gabapentin 12/20/2015 -30
1 Gabapentin 12/20/2015 -30
1 Gabapentin 12/20/2015 30
1 Gabapentin 12/20/2015 30
1 Gabapentin 12/20/2015 30
1 Gabapentin 12/20/2015 360
1 Gabapentin 12/21/2015 30
1 Gabapentin 12/21/2015 30
2 Naproxen 12/27/2016 -30
2 Naproxen 12/27/2016 30
2 Naproxen 12/27/2016 30
3 Ibuprofen 12/28/2016 -30
3 Ibuprofen 12/28/2016 -30
3 Ibuprofen 12/28/2016 30
3 Ibuprofen 12/28/2016 30
3 Ibuprofen 12/28/2016 30
4 Naproxen 12/29/2016 -30
4 Naproxen 12/29/2016 30
Next, we’re going to split the datasets into separate positive and negative datasets.
data positive;
set have (where = (qty_dspnd > 0)); /* #1 */
length count 3.; /* #2 */
by id drug service_dt; /* #3 */
if first.service_dt then count = 1; /* #4 */
else count + 1; /* #5 */
run;
data negative;
set have (where = (qty_dspnd < 0));
length count 3.;
by id drug service_dt;
if first.service_dt then count = 1;
else count + 1;
run;
Let’s back it up and go through the numbers I have up there.
- Here, we are subsetting our original dataset where the prescription quantity is positive.
- Note, this will be the same for the
negativedataset, but the direction will be reversed.
- Note, this will be the same for the
- For good measure and space consideration, set the length of the
countvariable to 3. - We are going to conduct by-group processing based on
id,drug, andservice_dt. - If it’s the first
service_dtfor a givenidordrug, then set ourcountvariable equal to 1. - Otherwise, if it isn’t the first
service_dtfor a givenidordrug, then incrementcountby 1.
Further, let’s see what the datasets look like. First, here’s the positive dataset:
id drug service_dt qty_dspnd count
1 Gabapentin 12/20/2015 30 1
1 Gabapentin 12/20/2015 30 2
1 Gabapentin 12/20/2015 360 3
1 Gabapentin 12/20/2015 30 4
1 Gabapentin 12/21/2015 30 1
1 Gabapentin 12/21/2015 30 2
2 Naproxen 12/27/2016 30 1
2 Naproxen 12/27/2016 30 2
3 Ibuprofen 12/28/2016 30 1
3 Ibuprofen 12/28/2016 30 2
3 Ibuprofen 12/28/2016 30 3
4 Naproxen 12/29/2016 30 1
And here’s the negative dataset:
id drug service_dt qty_dspnd count
1 Gabapentin 12/20/2015 -30 1
1 Gabapentin 12/20/2015 -30 2
2 Naproxen 12/27/2016 -30 1
3 Ibuprofen 12/28/2016 -30 1
3 Ibuprofen 12/28/2016 -30 2
4 Naproxen 12/29/2016 -30 1
Remember, the datasets were in ascending order by qty_dspnd before they were placed into separate positive and negative datasets. Accordingly, when we count the values, those with negative values will have the same count value as those with positve values. Any positive values that do not have a negative match will have an unmatched count value for a given id, drug, and service_dt.
Since we split the datasets by positive and negative values, we now need to merge them. We want to keep only those positive values that do not have a corresponding negative qty_dspnd value for a given id, drug, and service_dt.
data want
do_not_want;
merge positive (in = a rename = (qty_dspnd = qty_dspnd_pos)) /* #1 */
negative (in = b rename = (qty_dspnd = qty_dspnd_neg)); /* #2 */
by id drug service_dt count; /* #3 */
if a; /* #4 */
if sum(qty_dspnd_pos, qty_dspnd_neg) = 0 then output do_not_want; /* #5 */
else output want; /* #6 */
run;
Again, let’s go through what I have numbered up here:
- I create a
wantdataset to output the observations that I want to keep.- These are our positive values that do not have a corresponding negative
qty_dspndvalue.
- These are our positive values that do not have a corresponding negative
- I create a
do_not_wantdataset to output observations that I do not want to keep.- These are our negative values and positive
qty_dspndvalues that essentially zero each other out for a givenid,drug,service_dt, andcount.
- These are our negative values and positive
- I enter the first dataset that I want to
MERGE, namelypositive.- First, I use the
INoption to identify those records coming from thepositivedataset asarecords. If I remember the technical details correctly, this is a temporary variable that doesn’t get written to the dataset unless you explicitly request it. - Second, I rename the
qty_dspndvariable toqty_dspnd_pos. WithDATAstep merging, variables with the same name get overwritten in the PDV. In other words, the very last dataset that you merge will overwrite any existing values in the previous dataset(s) if they have the same name and type.
- First, I use the
- Same thing as above, just for the
negativedataset. - We then
MERGEthese two datasets byid,drug,service_dt, andcount. - Using the
INdata set option, I keep those records identified asarecords. This is essentially aLEFT JOINsince I want to keep records that have positive values only. - Here is where the magic happens. After merging by
id,drug,service_dt, andcount, if theqty_dspnd_posandqty_dspnd_negvalues sum to zero, then weOUTPUTthem to thedo_not_wantdataset. - Otherwise, if they do not sum to zero, we
OUTPUTthem to thewantdataset.

Let’s see the output. Here’s out want dataset:
id drug service_dt qty_dspnd_pos count qty_dspnd_neg
1 Gabapentin 12/20/2015 30 3 .
1 Gabapentin 12/20/2015 360 4 .
1 Gabapentin 12/21/2015 30 1 .
1 Gabapentin 12/21/2015 30 2 .
2 Naproxen 12/27/2016 30 2 .
3 Ibuprofen 12/28/2016 30 3 .
Here’s our do_not_want dataset:
id drug service_dt qty_dspnd_pos count qty_dspnd_neg
1 Gabapentin 12/20/2015 30 1 -30
1 Gabapentin 12/20/2015 30 2 -30
2 Naproxen 12/27/2016 30 1 -30
3 Ibuprofen 12/28/2016 30 1 -30
3 Ibuprofen 12/28/2016 30 2 -30
4 Naproxen 12/29/2016 30 1 -30
I could have done some cleanup and only kept the necessary variables, but I want you all to see exactly what happened.
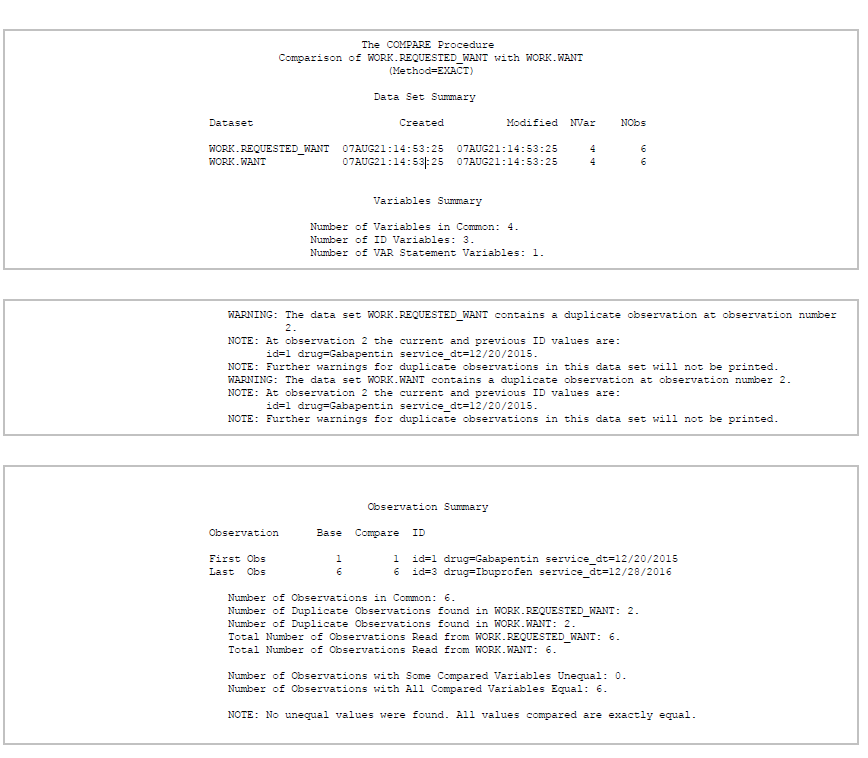
Just for verification purposes (and possibly hinting at another post), I’ll run PROC COMPARE, a procedure I used heavily at a previous job.
PROC COMPARE literally compares two datasets for equivalency. If the datasets have differing values, it gives you the location and values of those observations that do not match.
First, you have to use PROC SORT:
proc sort
data = want;
by id drug service_dt qty_dspnd_pos;
run;
proc sort
data = requested_want;
by id drug service_dt qty_dspnd;
run;
Now we use PROC COMPARE:
proc compare
listbase listcompare /* #1 */
base = requested_want /* #2 */
compare = want (keep = id drug service_dt qty_dspnd_pos rename = (qty_dspnd_pos = qty_dspnd)); /* #3 */
id id drug service_dt; /* #4 */
var qty_dspnd; /* #5 */
run;
Again, keeping this brief. This should be another post entirely:
- These options, I believe, will identify which values come from which dataset if any values differ.
- List the original dataset.
- List the compare dataset. I also restricted the variables and renamed the
qty_dspnd_posback toqty_dspnd.- It really doesn’t matter which one you label
baseorcompare–just remember which one is which.
- It really doesn’t matter which one you label
- Use an
IDstatement to identify which variables you want the comparison to be done on. - Finally, list the variable you want to compare.
It gives us this:
PROC COMPARE produces HTML output showing that our base and compare datasets are equivalent (see NOTE: No unequal values were found. All values compared are exactly equal. at the bottom). If you have a lot of differing values, then this output will be very long.
You could also do a PROC SQL join to determine equivalency, but again, I need to save this for another post.
As always, feel free to shoot me an email at michaelqmaguire2@cop.ufl.edu, message me on Teams, or stop by my office at HPNP 2330.