You ever had 10-45 diagnoses that you want to see, but they have some uninterpretable ICD/CPT code?
Maybe you have a large csv file that has every single ICD code and its respective CCS cross-walk.
In reality, you can have a dataset in your
WORKfolder that you can run through this same process. If you have it as a dataset, you can turn it into aFORMAT.
Someone had the following issue the other day:
1. They had up to 10 CPT codes per patient eyeing them down that didn't make any sense.
2. They had a large csv containing a CPT code range, a CCS code, and a CCS label.
3. They wanted the CCS label for each CPT code displayed.
Here is a mock-up of some patient-level data with some fake CPT codes for those that want to follow along:
data have_pt;
infile datalines delimiter = "," missover;
length enrolid $7. cpt1-cpt10 $5.;
input enrolid cpt1-cpt10;
datalines;
1234567,96020,G0255,96020,,96020,96020,NAAAH,64461
2234567,96105,64446,61150,C9729,COOL,22861
3322112,95999,63655,0169T,NOPE,NOPE,??CPT,IHATE,CPTCD,JUST,JOKIN
4221312,62267,28035,64461,62161,62164,62165
4222145,95965,0434T,0386T,63000,0109T,0437T
;
What it looked like after a PROC PRINT:
| enrolid | cpt1 | cpt2 | cpt3 | cpt4 | cpt5 | cpt6 | cpt7 | cpt8 | cpt9 | cpt10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1234567 | 96020 | G0255 | 96020 | 96020 | 96020 | NAAAH | 64461 | |||
| 2234567 | 96105 | 64446 | 61150 | C9729 | COOL | 22861 | ||||
| 3322112 | 95999 | 63655 | 0169T | NOPE | NOPE | ??CPT | IHATE | CPTCD | JUST | JOKIN |
| 4221312 | 62267 | 28035 | 64461 | 62161 | 62164 | 62165 | ||||
| 4222145 | 95965 | 0434T | 0386T | 63000 | 0109T | 0437T |
Here’s a portion of the CPT to CCS conversion we were looking at:
data have_ccs;
infile datalines delimiter = "," truncover;
input code_range :$15. ccs ccs_label $100.;
datalines;
'61000-61055',1,Incision and excision of CNS
'61105-61106',1,Incision and excision of CNS
'61108-61130',1,Incision and excision of CNS
'61150-61156',1,Incision and excision of CNS
'61250-61315',1,Incision and excision of CNS
'61320-61323',1,Incision and excision of CNS
'61340-61340',1,Incision and excision of CNS
'61345-61440',1,Incision and excision of CNS
'61470-61490',1,Incision and excision of CNS
'61510-61516',1,Incision and excision of CNS
'61518-61530',1,Incision and excision of CNS
'61534-61545',1,Incision and excision of CNS
'61556-61557',1,Incision and excision of CNS
'61570-61576',1,Incision and excision of CNS
'61582-61596',1,Incision and excision of CNS
'61598-61608',1,Incision and excision of CNS
'61615-61616',1,Incision and excision of CNS
'61712-61735',1,Incision and excision of CNS
'61880-61880',1,Incision and excision of CNS
'62161-62164',1,Incision and excision of CNS
'0169T-0169T',2,Insertion, replacement, or removal of extracranial ventricular shunt
'61107-61107',2,Insertion, replacement, or removal of extracranial ventricular shunt
'61210-61210',2,Insertion, replacement, or removal of extracranial ventricular shunt
'62160-62160',2,Insertion, replacement, or removal of extracranial ventricular shunt
'62180-62258',2,Insertion, replacement, or removal of extracranial ventricular shunt
'0202T-0202T',3,Laminectomy, excision intervertebral disc
'0274T-0275T',3,Laminectomy, excision intervertebral disc
'22856-22856',3,Laminectomy, excision intervertebral disc
'22858-22858',3,Laminectomy, excision intervertebral disc
'22861-22861',3,Laminectomy, excision intervertebral disc
'22864-22864',3,Laminectomy, excision intervertebral disc
'61343-61343',3,Laminectomy, excision intervertebral disc
'62287-62287',3,Laminectomy, excision intervertebral disc
'62351-62351',3,Laminectomy, excision intervertebral disc
'62380-63252',3,Laminectomy, excision intervertebral disc
'63655-63655',3,Laminectomy, excision intervertebral disc
'63709-63709',3,Laminectomy, excision intervertebral disc
'63740-63740',3,Laminectomy, excision intervertebral disc
'C9729-C9729',3,Laminectomy, excision intervertebral disc
'G0276-G0276',3,Laminectomy, excision intervertebral disc
'S2348-S2348',3,Laminectomy, excision intervertebral disc
'S2350-S2350',3,Laminectomy, excision intervertebral disc
'S2351-S2351',3,Laminectomy, excision intervertebral disc
'62270-62272',4,Diagnostic spinal tap
'62263-62264',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'62274-62282',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'62288-62289',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'62298-62298',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'62310-62350',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'62355-62355',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'63600-63610',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'63650-63650',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'63657-63688',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'63750-63780',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'64412-64412',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'64416-64416',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'64446-64446',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'64448-64449',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'64461-64484',5,Insertion of catheter or spinal stimulator and injection into spinal canal
'28035-28035',6,Decompression peripheral nerve
'29848-29848',6,Decompression peripheral nerve
'64702-64727',6,Decompression peripheral nerve
'0077T-0077T',7,Other diagnostic nervous system procedures
'0106T-0110T',7,Other diagnostic nervous system procedures
'0199T-0199T',7,Other diagnostic nervous system procedures
'0285T-0285T',7,Other diagnostic nervous system procedures
'0333T-0333T',7,Other diagnostic nervous system procedures
'0341T-0341T',7,Other diagnostic nervous system procedures
'0381T-0386T',7,Other diagnostic nervous system procedures
'0434T-0436T',7,Other diagnostic nervous system procedures
'61140-61140',7,Other diagnostic nervous system procedures
'61750-61751',7,Other diagnostic nervous system procedures
'61795-61795',7,Other diagnostic nervous system procedures
'62267-62267',7,Other diagnostic nervous system procedures
'62269-62269',7,Other diagnostic nervous system procedures
'63690-63690',7,Other diagnostic nervous system procedures
'64795-64795',7,Other diagnostic nervous system procedures
'92516-92516',7,Other diagnostic nervous system procedures
'95857-95872',7,Other diagnostic nervous system procedures
'95874-95882',7,Other diagnostic nervous system procedures
'95885-95943',7,Other diagnostic nervous system procedures
'95965-95982',7,Other diagnostic nervous system procedures
'95999-96003',7,Other diagnostic nervous system procedures
'96020-96020',7,Other diagnostic nervous system procedures
'96105-96115',7,Other diagnostic nervous system procedures
'G0255-G0255',7,Other diagnostic nervous system procedures
'G0453-G0453',7,Other diagnostic nervous system procedures
'S3900-S3900',7,Other diagnostic nervous system procedures
'S3905-S3905',7,Other diagnostic nervous system procedures
'S8040-S8040',7,Other diagnostic nervous system procedures
;
This is what 10 of those observations looked like after a PROC PRINT:
| code_range | ccs | ccs_label |
|---|---|---|
| ‘61000-61055’ | 1 | Incision and excision of CNS |
| ‘61105-61106’ | 1 | Incision and excision of CNS |
| ‘61108-61130’ | 1 | Incision and excision of CNS |
| ‘61150-61156’ | 1 | Incision and excision of CNS |
| ‘61250-61315’ | 1 | Incision and excision of CNS |
| ‘61320-61323’ | 1 | Incision and excision of CNS |
| ‘61340-61340’ | 1 | Incision and excision of CNS |
| ‘61345-61440’ | 1 | Incision and excision of CNS |
| ‘61470-61490’ | 1 | Incision and excision of CNS |
| ‘61510-61516’ | 1 | Incision and excision of CNS |
So, we wanted those CPT codes in have_pt to be displayed with the according CCS labels in have_ccs.

One of the best tools, in my opinion, for this kind of task is PROC FORMAT.
PROC FORMAT has an option called CNTLIN =. This can save you lots of time in scenarios where you have some large, existing dataset with formats already written for you.
First, though, we need to prepare the data set for PROC FORMAT.
The code to get this is a bit peculiar and esoteric – just know that. There’s nothing you can do but learn this weird code. It makes sense once you do it enough.
Let me highlight the necessary elements before I throw the code at you:
You need a
startvariable representing the starting value for the label/format.
You need an
endvariable representing the ending value for the label/format.
You need a
labelvariable representing, you guessed it, the label.
You need a
fmtnamevariable representing the name of the format.
You need a
typevariable representing the kind of format you want (e.g., ‘c’ for character, ‘n’ for numeric).
You need an
hlovaraiable representing the ‘high-low-other’ parameters that you can use in a regularPROC FORMATrequest.
You also need to add the
END =option on yourSETstatement so you can add an additional observation at the end of the dataset. You’ll see what I mean later.
data have_fmt (drop = code_range); /* #1 */
set have_ccs (
keep = code_range ccs_label /* #2(a) */
rename = (ccs_label = label) /* #2(b) */
)
end = z; /* #2(c) */
retain fmtname "$cpt_fmt" type "c"; /* #3 */
start = scan(compress(code_range, "'"), 1, "-"); /* #4 */
end = scan(compress(code_range, "'"), -1, "-"); /* #5 */
output; /* #6 */
if z then do; /* #7 */
start = ""; /* #7(a) */
end = ""; /* #7(b) */
label = "Missing"; /* #7(c) */
hlo = "O"; /* #7(d) */
output; /* #7(e) */
end; /* #7(f) */
run; /* #8 */
Woohoo, lots of fun, step-by-step explanations for me!
-
Our
DATAstep is a usualDATAstep. For those that don’t know, including aDROP =on yourDATAstatement drops variables at the very, very end of processing, before it’s written to the dataset. -
Our
SETstatement does a couple things. Similiar to (1), anyKEEP,DROP,RENAMEthat you conduct on yourSETstatement affects how the data is read into SAS. It usually speeds up processing. Just know thatDROP,KEEP, andRENAMEoptions on theDATAstep happen at the end, whileDROP,KEEP, andRENAMEoptions happen at the beginning of processing.a. We are keeping
code_rangeandccs_labelfrom the input data set. Since this is on theSETstatement, these are the only variables read in.b. I’m also going to
RENAMEccs_labeltolabel, which is required forPROC FORMAT. I really meant that this code was esoteric.c. Notice that I used the
END =option before I insert a semicolon to end theSETstatement. This creates, I believe, a temporary variable in the PDV designating whether a record in the data set is the last observation in the data set. If it is, it get’s assigned the value “z”. You could make it anything, though (e.g., a, b, c, gator). -
We use
RETAINto create a variable namefmtname, and we set the value forfmtnameto be equal to$cpt_fmt. We also create a variable namedtypeand set the value toc, which standsd for character. As an aside,RETAINcan be used to create static variables across a data set, and it’s much faster than explicitly creating it likefmtname = "$cpt_fmt";.a. I think the
$is unnecessary, but I like to put it in there for good measure. -
Here, I’m cleaning to get the
startvalue for the format.i. First, I’m using
SCANto extract the value that I want at a given location.ii. Second, I’m using
COMPRESSto remove those pesky quotes surrounding thecode_rangevalue. Note, this is my first argument forSCAN, so we’re essentially going toSCANthe unquoted version ofcode_range.iii.
SCANrequires a position argument for you to extract your value of interest. Here, I want to take the first value (1), and I want it to be the first value before the hyphen (-). -
Literally the exact same as (4), except I’m getting the
endvalue. The only difference is my positional argument inSCAN, which I change to-1. Specifying a-1takes the outermost number (think right to left) before the hyphen (-). -
Now we add an explicit
OUTPUTstatement. In SAS, there is an implicitOUTPUTstatement at the end of aDATAstep/when an observation has been fully processed in the PDV. Using an explicitOUTPUTstatement overrides the implicitOUTPUT. So here, we’re usingOUTPUTto send each observation that is read in and cleaned to the data sethave_fmtonce it has been cleaned. -
Here’s that
ENDoption that we talked about in 2(c) comes into play. We’re basically sayingifthis is the last observation (z),then dothe following:a. For the
startvariable, set it to missing ("").b. For the
endvariable mentioned in (5), set it to missing (""). This is not theEND =option.c. For the
labelvariable, I’m going to set it toMissingso that it’s helpful to see when something isn’t coded correctly.d. Now, here’s that
hlovariable. I set it toOto represent “other” codes. Basically, any CPT code in thehave_ptthat does not have a corresponding CCS code inhave_ccswill have a missing label ("").e. We want to
OUTPUTthis record to the dataset AFTER all the other codes that were read in.f.
ENDthatif-then-doprocessing. -

Weird, huh?
Let’s see what the first ten observations look like:
| label | fmtname | type | start | end | hlo |
|---|---|---|---|---|---|
| Incision and excision of CNS | $cpt_fmt | c | 61000 | 61055 | |
| Incision and excision of CNS | $cpt_fmt | c | 61105 | 61106 | |
| Incision and excision of CNS | $cpt_fmt | c | 61108 | 61130 | |
| Incision and excision of CNS | $cpt_fmt | c | 61150 | 61156 | |
| Incision and excision of CNS | $cpt_fmt | c | 61250 | 61315 | |
| Incision and excision of CNS | $cpt_fmt | c | 61320 | 61323 | |
| Incision and excision of CNS | $cpt_fmt | c | 61340 | 61340 | |
| Incision and excision of CNS | $cpt_fmt | c | 61345 | 61440 | |
| Incision and excision of CNS | $cpt_fmt | c | 61470 | 61490 | |
| Incision and excision of CNS | $cpt_fmt | c | 61510 | 61516 |
Let’s look at the last observations of that just to see what happened with all that hlo stuff:
| label | fmtname | type | start | end | hlo |
|---|---|---|---|---|---|
| Other diagnostic nervous system procedures | $cpt_fmt | c | S8040 | S8040 | |
| Missing | $cpt_fmt | c | O |
See how the if z then do outputted an observation to the bottom of the dataset? The record right above the hlo = "O" was marked internally as z with our end = z option. Once we explictly outputted that record, SAS encountered the Z and concatenated an additional record with our specifications.
If you’re kind of lost with the above DATA step and prefer a PROC SQL alternative, you can do this:
proc sql;
create table have_fmt_sql as
select
scan(compress(code_range,("'")), 1, "-") as start,
scan(compress(code_range,("'")), -1, "-") as end,
ccs_label as label,
"$cpt_fmt" as fmtname,
"c" as type,
"" as hlo
from
have_ccs;
insert into have_fmt_sql
values ("", "", "Missing", "$cpt_fmt", "c", "O");
quit;
They provide the same results, but the DATA step version requires a bit more manipulation of the PDV, as well as some weird options that you don’t always use. The PROC SQL version ends up looking a little cleaner, in my opinion.
Okay, now we’re ready to throw it in PROC FORMAT.

Take either the DATA step version or the PROC SQL version and use it as your argument for CNTLIN:
proc format
cntlin = have_fmt;
run;
That was a bit anticlimactic. This is what your log should show, though:
138 proc format
139 cntlin = have_fmt;
NOTE: Format $CPT_FMT has been output.
140 run;
Alright, so now we have converted have_ccs from a data set into a FORMAT. As most of you probably know, applying a FORMAT is pretty easy.
Let’s say that we want to see the variables themselves in the dataset.
data have_pt_w_fmt_vars (drop = i); /* #1 */
set have_pt; /* #2 */
array _cpt [*] cpt:; /* #3 */
array _fcpt [10] $100. _fcpt1 - _fcpt10; /* #4 */
do i = 1 to dim(_cpt); /* #5 */
_fcpt[i] = put(_cpt[i], $cpt_fmt.); /* #5(a) */
end; /* #5(b) */
run;

-
Create a new data set. Drop
iright at the end. -
Set our
have_ptdata set. -
Create an
ARRAYthat groups together all thecptvariables (cpt1 - cpt10). The colon is a modifier that tells SAS to include all variables starting withcptto be included in this array or group. -
Create another
ARRAYthat creates ten variables, _fcpt1 - _fcpt10 (meaning formatted CPT). I’m also setting the length of these character variables to $100. -
Do the following from the first instance of the
_cptarray to the last instance.DIMessentially sets the upper bound of the array for you.a. The
DOloop will iterate from 1 to 10 here. So the first instance will do the following:- _fcpt1 = put(cpt1, $cpt_fmt.);b. End the
DOloop and incrementiby 1. It will return to the top of the do loop and repeat (e.g., next instance will be 2) until theivariable exceeds the upper bound of the array (Here,iwill become 11. You can comment out thedrop = ifrom theDATAstep above to see).
This creates ten new variables, and each one can accomodate a format of up to 100 characters. It’s also good to cross-walk them this way to ensure you mapped on the formats correctly, but it can take up a lot of space depending on the number of variables. I recommend some other ways below so that you don’t have to permanently alter the values within your data set.
| enrolid | cpt1 | cpt2 | _fcpt1 | _fcpt2 |
|---|---|---|---|---|
| 1234567 | 96020 | G0255 | Other diagnostic nervous system procedures | Other diagnostic nervous system procedures |
| 2234567 | 96105 | 64446 | Other diagnostic nervous system procedures | Insertion of catheter or spinal stimulator and injection into spinal canal |
| 3322112 | 95999 | 63655 | Other diagnostic nervous system procedures | Laminectomy, excision intervertebral disc |
| 4221312 | 62267 | 28035 | Other diagnostic nervous system procedures | Decompression peripheral nerve |
| 4222145 | 95965 | 0434T | Other diagnostic nervous system procedures | Other diagnostic nervous system procedures |
Further, we can see that those with incorrect CPT codes didn’t get mapped.
| enrolid | cpt6 | cpt7 | cpt8 | _fcpt6 | _fcpt7 | _fcpt8 |
|---|---|---|---|---|---|---|
| 1234567 | NAAAH | 64461 | Insertion of catheter or spinal stimulator and injection into spinal canal | |||
| 2234567 | 22861 | Laminectomy, excision intervertebral disc | ||||
| 3322112 | ??CPT | IHATE | CPTCD | |||
| 4221312 | 62165 | |||||
| 4222145 | 0437T |
We can, of course, apply the FORMAT directly to the dataset variables, but it permanently alters them. You could accomplish that by doing this:
data permanent;
set have_pt;
format cpt: $cpt_fmt.;
run;
A lot less code, but you’ve overwritten everything and now you have a lot of long strings instead of those short codes.
You can always just PROC PRINT to check a couple observations. Here, I format the first PROC PRINT, but leave the second untouched.
proc print
data = have_pt (obs = 10);
format cpt: $cpt_fmt.;
run;
proc print
data = have_pt (obs = 10);
run;
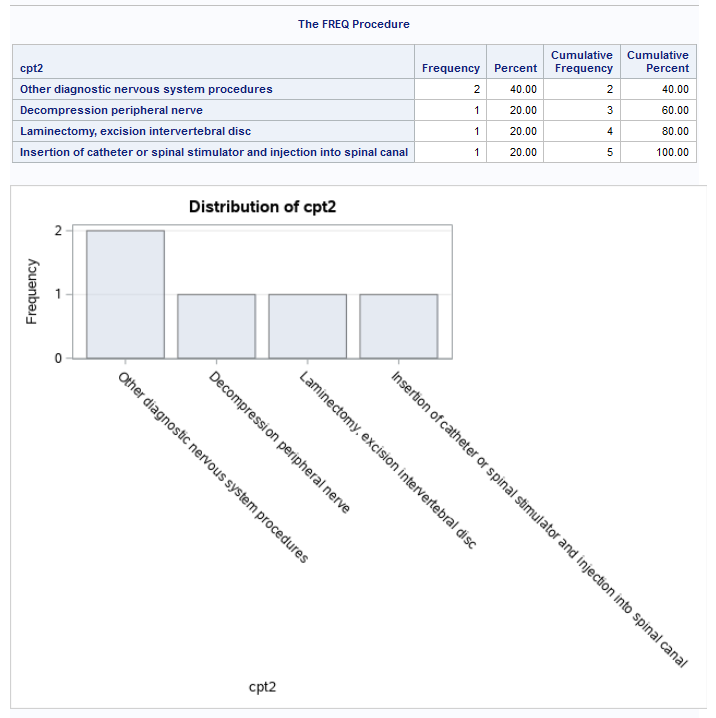
But you can apply that format everywhere. We could do a PROC FREQ with the PLOTS = FREQPLOT option and apply the format. Let’s look at cpt2.
proc freq
data = have_pt;
tables cpt2 / plots = freqplot;
format cpt2 $cpt_fmt.;
run;
FYI: you can specify
cpt1 - cpt10(orcpt:) on theTABLESstatement to get all of them printed out with a frequency plot. You need to adjust the format toFORMATtoformat cpt1-cpt10 $cpt_fmt.(orcpt:).
Nonetheless, our frequency plot of cpt2 shows this:
Finally, let’s say we want to see the most common CPT codes cross-walked with their respective CCS labels.
First, I need to transpose the data:
proc transpose
data = have_pt
out = have_pt_tp (
rename = (
col1 = cpt_code
_name_ = cpt_position
)
);
by enrolid;
var cpt:;
run;
This gives us this. This is truncated to ten observations:
| enrolid | cpt_position | cpt_code |
|---|---|---|
| 1234567 | cpt1 | 96020 |
| 1234567 | cpt2 | G0255 |
| 1234567 | cpt3 | 96020 |
| 1234567 | cpt4 | 96020 |
| 1234567 | cpt5 | 96020 |
| 1234567 | cpt6 | NAAAH |
| 1234567 | cpt7 | 64461 |
| 1234567 | cpt8 | |
| 1234567 | cpt9 | |
| 1234567 | cpt10 | |
| 2234567 | cpt1 | 96105 |
| 2234567 | cpt2 | 64446 |
| 2234567 | cpt3 | 61150 |
| 2234567 | cpt4 | C9729 |
| 2234567 | cpt5 | COOL |
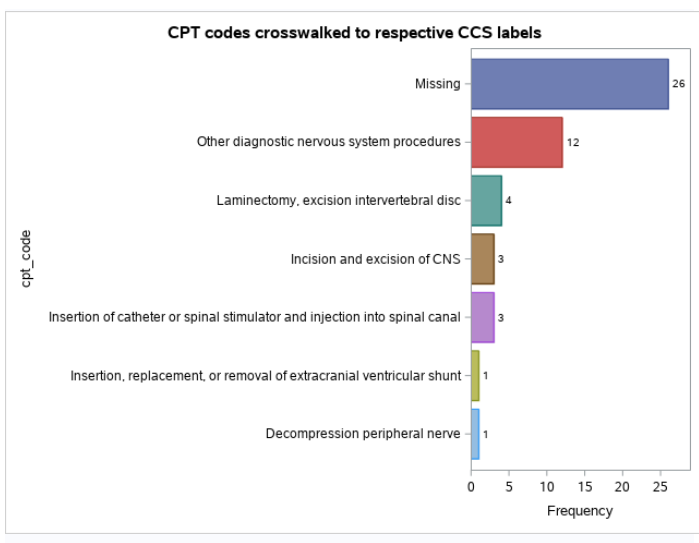
Now I’m going to plot it with PROC SGPLOT.
Pardon my weird formatting - I find that SAS graphing commands/statements are awkward with spacing and difficult to follow.
I tend to add tabs and carriage returns to normalize things. I also add semicolons below the statement to denote exactly where a statement ends intstead of searching for it at the end of a statement.
title "CPT codes crosswalked to respective CCS labels";
proc sgplot
data = have_pt_tp
;
hbar cpt_code / stat = freq
group = cpt_code
datalabel
missing
categoryorder = respdesc
;
format cpt_code $cpt_fmt.
;
run;
title;
And here is that plot.
I hope this was a helpful resource for you all.
If you have any questions or run into any issues, please let me know and I will address them as soon as possible.
The program for this post can be downloaded here.